Basic structure calculation starting from given restraints: Difference between revisions
No edit summary |
No edit summary |
||
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
This calculation shows how to calculate a structure from previously assigned upper limits. The input data are in the subdirectory | This calculation shows how to calculate a structure from previously assigned upper limits and dihedral angle restraints. The input data are in the 'demo/basic' subdirectory of the CYANA software package: | ||
;demo.seq:amino acid sequence | ;demo.seq:amino acid sequence | ||
;demo.upl:upper distance limit restraints | ;demo.upl:upper distance limit restraints | ||
;demo.aco:dihedral angle restraints | ;demo.aco:dihedral angle restraints | ||
;CALC.cya:macro for the structure calculation | ;CALC.cya:macro for the structure calculation | ||
When CYANA is started, it automatically reads the standard amino acid library and the amino acid sequence of the protein (if multiple sequence files are in the current directory, the most recently modified one is read). | |||
When CYANA is started, it automatically | |||
The actual structure calculation is performed by the commands in the macro file '''CALC.cya''': | The actual structure calculation is performed by the commands in the macro file '''CALC.cya''': | ||
Latest revision as of 08:27, 28 July 2019
This calculation shows how to calculate a structure from previously assigned upper limits and dihedral angle restraints. The input data are in the 'demo/basic' subdirectory of the CYANA software package:
- demo.seq
- amino acid sequence
- demo.upl
- upper distance limit restraints
- demo.aco
- dihedral angle restraints
- CALC.cya
- macro for the structure calculation
When CYANA is started, it automatically reads the standard amino acid library and the amino acid sequence of the protein (if multiple sequence files are in the current directory, the most recently modified one is read).
The actual structure calculation is performed by the commands in the macro file CALC.cya:
read upl demo.upl # read upper distance bounds read aco demo.aco # read dihedral angle restraints calc_all 50 steps=4000 # calculate conformers overview demo.ovw structures=10 pdb # analyze and write best conformers
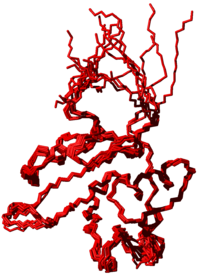
The first two commands read the input files with upper distance bounds and dihedral angle restraints, respectively. Then, 50 conformers are calculated using the standard simulated annealing schedule with 4000 torsion angle dynamics steps per conformer. Finally, the 10 conformers with the lowest final target function values are analyzed. The coordinates of these 10 best conformers are written to the PDB file demo.pdb, and an overview table is saved in the file demo.ovw:
Structural statistics:
str target upper limits van der Waals torsion angles
function # rms max # sum max # rms max
1 1.56 1 0.0073 0.36 5 5.2 0.34 0 0.3460 3.19
2 1.87 2 0.0084 0.36 5 6.1 0.33 0 0.3813 3.63
3 1.92 2 0.0083 0.36 5 5.8 0.36 0 0.3491 3.29
4 1.94 1 0.0082 0.36 6 6.4 0.34 0 0.3478 3.46
5 2.02 2 0.0080 0.35 7 5.7 0.34 0 0.5790 4.35
6 2.05 1 0.0082 0.35 8 6.0 0.34 0 0.5704 4.29
7 2.12 2 0.0092 0.36 4 6.5 0.35 0 0.3629 3.47
8 2.20 4 0.0105 0.36 5 7.2 0.31 0 0.3597 3.45
9 2.24 2 0.0087 0.36 6 7.4 0.34 0 0.3843 3.47
10 2.34 3 0.0119 0.61 5 6.6 0.35 0 0.3309 3.05
Ave 2.03 2 0.0089 0.38 6 6.3 0.34 0 0.4011 3.56
+/- 0.21 1 0.0013 0.08 1 0.7 0.01 0 0.0881 0.41
Min 1.56 1 0.0073 0.35 4 5.2 0.31 0 0.3309 3.05
Max 2.34 4 0.0119 0.61 8 7.4 0.36 0 0.5790 4.35
Cut 0.20 0.20 5.00
Constraints violated in 3 or more structures:
# mean max. 1 5 10
Upper HA ASP 68 - H ASN 69 3.28 4 0.11 0.29 +* ++ peak 1162
Upper HB ILE 85 - H ASP 86 3.80 10 0.36 0.36 +++++++++* peak 803
VdW N LEU 39 - CD1 LEU 39 3.05 10 0.23 0.24 ++++++++*+
VdW N ILE 81 - CD PRO 82 3.05 10 0.26 0.28 ++*+++++++
VdW N ILE 81 - HD2 PRO 82 2.45 10 0.34 0.36 ++*+++++++
VdW CG2 ILE 81 - C ILE 81 2.90 6 0.20 0.22 ++ ++ *+
VdW CB THR 91 - H GLN 92 2.55 7 0.22 0.27 ++++++ *
VdW O THR 91 - CB GLN 92 2.90 3 0.15 0.20 * ++
VdW HA VAL 107 - CD PRO 108 2.60 4 0.18 0.30 *+ + +
2 violated distance restraints.
7 violated van der Waals restraints.
0 violated angle restraints.
RMSDs for residues 10..100:
Average backbone RMSD to mean : 0.52 +/- 0.09 A (0.40..0.72 A; 10 structures)
Average heavy atom RMSD to mean : 1.04 +/- 0.06 A (0.96..1.17 A; 10 structures)