Automated resonance assignment with FLYA (Gothenburg 2021)
In this tutorial we will determine the resonance assignments and the structure of a protein using the program CYANA.
Installation of CYANA demo version
If not done yet, please install the demo version of CYANA.
Experimental input data
Example data for FLYA is in the 'demo/flya' directory of the CYANA package.
The protein sequence is stored in three-letter code in the file 'demo.seq'.
Experimental peak lists are available for the following spectra:
- [1H,13C]-HSQC (called 'C13HSQC' in FLYA)
- [1H,15N]-HSQC (called 'N15HSQC' in FLYA)
- 3D [13C]-resolved NOESY (called 'C13NOESY' in FLYA)
- 3D [15N]-resolved NOESY (called 'N15NOESY' in FLYA)
- HNCA
- HN(CO)CA (called 'HNcoCA' in FLYA)
- HNCO
- HN(CA)CO (called 'HNcaCO' in FLYA)
- CBCANH
- CBCACONH (called 'CBCAcoNH' in FLYA)
- HBHACONH (called 'HBHAcoNH' in FLYA)
- HCCH-TOCSY (called 'HCCHTOCSY' in FLYA)
- HCCH-COSY (called 'HCCHCOSY' in FLYA)
- C(CO)NH (called 'CcoNH' in FLYA)
- HC(CO)NH (called 'HCcoNH' in FLYA)
Peak lists in XEASY format that have been prepared by automatic peak picking with the program NMRView are stored in files XXX.peaks, where XXX denotes the FLYA spectrum type.
Each peak list starts with a header that defines the experiment type and the order of dimensions. For instance, for HNCA.peaks:
# Number of dimensions 3
#FORMAT xeasy3D
#INAME 1 HN
#INAME 2 C
#INAME 3 N
#SPECTRUM HNCA HN C N
5 6.475 58.033 98.548 1 U 2.769E+02 0.000E+00 e 0 0 0 0
6 6.476 62.123 98.126 1 U 2.571E+01 0.000E+00 e 0 0 0 0
7 6.475 54.017 98.159 1 U 2.547E+01 0.000E+00 e 0 0 0 0
The first line specifies the number of dimensions (3 in this case). The next 4 lines ('#FORMAT' and '#INAME') are ignored by CYANA. The '#SPECTRUM' line is crucial and gives the experiment type (HNCA, which refers to the corresponding experiment definition in the CYANA library), followed by an identifier for each dimension of the peak list (HN C N) that specifies which chemical shift is stored in the corresponding dimension of the peak list. These labels must match those in the corresponding experiment definition in the general CYANA library (see below). After the '#SPECTRUM' line follows one line for every peak. For example, the first peak in the 'HNCA.peaks' list has
- Peak number 5
- HN chemical shift 6.475 ppm
- C (i.e. CA) chemical shift 58.033 ppm
- N chemical shift 98.548 ppm
The other data are irrelevant for automated chemical shift assignment with FLYA. In particular, the peak volume or intensity (2.769E+02) is not used by the algorithm.
Hint: The formats of other CYANA files are described in the CYANA Reference Manual.
Experiment definitions in the CYANA library
When you start CYANA, the program reads the library and displays the full path name of the library file. You can open the standard library file to inspect, for example, the NMR experiment definitions that define how expected peaks are generated by FLYA. For instance, the definition for the HNCA spectrum (search for 'HNCA' in the library file 'cyana.lib') is
SPECTRUM HNCA HN N C 0.980 HN:H_AMI N:N_AM* C:C_ALI C_BYL 0.800 HN:H_AMI N:N_AMI (C_ALI) C_BYL C:C_ALI
The first line corresponds to the '#SPECTRUM' line in the peak list. It specifies the experiment name and a label for the atoms that are detected in each dimension of the spectrum. The number of labels defines the dimensionality of the experiment (3 in case of HNCA).
Each line below defines a (formal) magnetization transfer pathway that gives rise to an expected peak. in the case of HNCA there are two lines, corresponding to the intraresidual and sequential peak. For instance, the definition for the intraresidual peak starts with the probability to observe the peak (0.980), followed by a series of atom types, e.g. H_AMI for amide proton etc. An expected peak is generated for each molecular fragment in which these atom types occur connected by single covalent bonds. The atoms whose chemical shifts appear in the spectrum are identified by their labels followed by ':', e.g. for HNCA 'HN:', 'N:', and 'C:'. The additional atom types refer to atoms that are not detected but must be present in a matching molecular fragment. An atom type in parenthesis indicates a branch in the molecular fragment. For instance, in the second magnetization transfer pathway that specifies the sequential HNCA peak, '(C_ALI)' indicates that the atom 'N:N_ALI' must be connected by a covalent bond to both a C_ALI (i.e. CA) and a C_BYL (i.e. C' of the preceding residue.
FLYA execution scripts
The CYANA scripts ("macros") 'CALC*.cya' contain the commands to perform various automated chemical shift assignment calculations.
For instance, 'CALCbackbone.cya' performs automated backbone resonance assignment. It starts with the specification of the names of the input peak lists:
peaks:=N15HSQC,HNCA,HNcaCO,HNCO,HNcoCA,CBCANH,CBCAcoNH
The peak list names are separated by commas (without blanks!). The files on disk have the file name extension .peaks, e.g. HNCA.peaks.
The commands above will use all available peak lists. You can choose any subset of them by modifying the 'peaks:=...' statement.
These are followed by tolerances for chemical shift matching:
assigncs_accH=0.03 assigncs_accC=0.4 assigncs_accN=assigncs_accC tolerance:=$assigncs_accH,$assigncs_accH,$assigncs_accC
In this case, a tolerance of 0.03 ppm will be used for protons, and 0.4 ppm for carbon and nitrogen.
The next parameter specifies the seed value for the random number generator (an arbitrary positive integer is ok).
randomseed=101
Groups of atoms for which assignment statistics will be calculated and reported in the 'flya.txt' output file can be defined like this:
analyzeassign_group := BB: N H CA CB C
The next commands restrict the generation of expected peaks to a subset of atoms, here the backbone atoms:
command select_atoms atom select "N H CA CB C" end
In this case, the command defines a group called BB (a name that can be chosen freely) comprising the atoms N, H, CA, CB, C.
Specific labeling can be handled in the same way. Peak list-specific atom selections can be applied as follows (not used in 'CALCbackbone.cya' but in 'CALClabeling.cya'):
command XXX_select atoms select "..." end
If desired, a "quick" optimization schedule can be used in the FLYA examples in order to speed up the calculation by inserting the following line above the 'flya ...' command:
shiftassign_quick=1
In production runs, better results can be expected (at the expense of longer computation times) if this variable is not set. Finally, there is the command to start the FLYA algorithm:
flya runs=10 assignpeaks=$peaks structure= shiftreference=ref.prot
Here, the given parameters of the 'flya' command specify that
- The number of independent runs of the algorithm, from which the consolidated shift will be calculated (chosen smaller than in normal production runs in order to speed up the calculation).
- The input peak lists that will be used (as defined above).
- No ensemble of random structures will be calculated for generating expected peaks (is only necessary for NOESY-type experiments).
- The results will be compared with the reference chemical shifts in the file 'ref.prot' (which have been determined independently by conventional methods). The reference chemical shifts will not be used by the algorithm but only for a subsequent analysis of its results.
Run a FLYA calculation
To run a FLYA calculation, you start CYANA and execute the corresponding 'CALC*.cya' script. For instance:
cyana "nproc=10; CALCbackbone"
By specifying 'nproc=10', 10 independent runs of the algorithm will be performed in parallel. On a computer with multiple processors this will speed up the calculation, which is expected to take a few minutes. For FLYA, the value of 'nproc' should correspond to the number of independent FLYA runs, i.e. the 'runs=10' parameter of the above 'flya' command.
FLYA output files
The FLYA algorithm will produce the following output files:
- flya.prot: Consensus assigned chemical shifts. This file contains a chemical shift for every atom that has been assigned to least one peak.
- flya.tab: Table with details about the chemical shift assignment of each atom (comparison with reference shifts). In this file you can see for each atom whether the assignment is "strong" (self-consistent) or "weak" (only tentative).
- flya.txt: Assignment statistics
- flya.pdf: Graphical representation of the assignment results
- XXX_exp.peaks: List of expected peaks, corresponding to input peak list XXX.peaks
- XXX_asn.peaks: Assigned peak list, corresponding to input peak list XXX.peaks
The flya.txt file
This output file starts with overall assignment statistics for each group of atoms as defined by 'analyzeassign_group:=...' in CALCbackbone.cya':
____________________________________________________________ CHEMICAL SHIFT ASSIGNMENT ____________________________________________________________ SEED: 1 chemical shifts for 542 atoms found Peaks assigned from frequencies BB: REFERENCES(2):512 CHEMICALSHIFTS(1):542 (1)and(2):512 MATCH:507(99.0% of (2))
- REFERENCES(2) is the number of reference assignments (in the selected group)
- CHEMICALSHIFTS(1) is is the number of atoms assigned by FLYA
- (1)and(2) is the number of atoms that are assigned by FLYA and in the reference.
- MATCH is the number of atoms with the same assignment by FLYA and in the reference. The percentage is relative to the number of reference assignments.
Further below comes a table with information about each peak list:
PEAKLISTS #Expected: Total number of expected peaks noRef: Number of expected peaks with missing reference shifts noPeak: Number of expected peaks for wich no peak can be measured Assigned: Number of expected peaks that could be assigned Match: Number of assigned peaks that fit reference shifts #Measured: Total number of peaks in peak list Assigned: Number of measured peaks that could be assigned to expected peaks exp/meas: Ratio of assigned expected and measured peaks Lists #Expected noRef noPeak Assigned Match #Measured Assigned exp/meas Assigned N15HSQC 106 8 1 104( 98.11%) 97( 91.51%) 131 96( 73.28%) 1.1 HNCA 211 15 11 194( 91.94%) 186( 88.15%) 329 179( 54.41%) 1.1 HNcaCO 211 15 11 197( 93.36%) 183( 86.73%) 246 176( 71.54%) 1.1 HNCO 105 7 1 101( 96.19%) 97( 92.38%) 158 97( 61.39%) 1.0 HNcoCA 105 7 0 101( 96.19%) 97( 92.38%) 158 99( 62.66%) 1.0 CBCANH 399 26 25 361( 90.48%) 350( 87.72%) 623 339( 54.41%) 1.1 CBCAcoNH 200 13 2 196( 98.00%) 185( 92.50%) 324 192( 59.26%) 1.0 ALL 1337 91 51 1254( 93.79%) 1195( 89.38%) 1969 1178( 59.83%) 1.1
It contains the following data:
- #Expected: Total number of expected peaks
- noRef: Number of expected peaks with missing reference shifts
- noPeak: Number of expected peaks for which no peak can be measured
- Assigned: Number of expected peaks that could be assigned based on the reference chemical shift assignments. The theoretical maximum of 100% corresponds to the situation that the spectra “explain” all expected peaks. Each expected peak can be mapped to at most one measured peak. Remaining expected peaks correspond to missing peaks in the measured peak list.
- Match: Number of assigned peaks that fit (within tolerance) reference shifts. The theoretical maximum of 100% corresponds to having all measured peaks assigned. Note that several expected peaks can be mapped to the same measured peak, i.e. the assignments of measured peaks can be unambiguous or ambiguous. Remaining unassigned measured peaks are likely to be artifacts.
- #Measured: Total number of peaks in peak list
- Assigned: Number of measured peaks that could be assigned to expected peaks
- exp/meas: Ratio of assigned expected and measured peaks
There is more information on the results of the assignment calculation in the 'flya.txt' file (not described here).
The flya.tab file
This file provides information about the chemical shift assignment of each individual atom:
Atom Residue Ref Shift Dev Extent inside inref ... N GLY 57 102.109 102.043 0.066 10.0 100.0 100.0 strong= H GLY 57 8.571 8.570 0.001 10.0 100.0 100.0 strong= CA GLY 57 45.415 45.433 -0.018 10.0 100.0 100.0 strong= HA2 GLY 57 4.042 HA3 GLY 57 3.436 C GLY 57 173.621 173.662 -0.041 10.0 89.4 90.0 strong= N LEU 58 120.640 120.649 -0.009 10.0 80.0 80.0 = H LEU 58 7.488 7.492 -0.004 10.0 79.8 80.0 = CA LEU 58 51.943 51.940 0.003 10.0 70.0 70.0 = HA LEU 58 4.995 CB LEU 58 45.602 45.568 0.034 10.0 82.7 80.0 strong= CG LEU 58 26.528 HG LEU 58 1.515 CD1 LEU 58 24.745 C LEU 58 173.619 174.576 -0.957 10.0 40.1 10.0 ! (C 59) ...
- Ref: Chemical shift value in the reference chemical shift list (ref.prot). It was not used in the calculation.
- Shift: Consensus chemical shift value from FLYA
- Dev = Ref - Shift
- Extent: Number of runs in which the atom was assigned by FLYA.
- Inside: Percentage of chemical shift values from the (10) independent runs of FLYA that agree (within the tolerance) with the consensus value.
- inref: Percentage of chemical shift values from the (10) independent runs of FLYA that agree (within the tolerance) with the reference value.
- Outcome of the assignment:
- strong: "strong" assignment, i.e. Inside > 80%.
- =: Assignment that agrees with reference, i.e. Dev < tolerance.
- !: Assignment that does not agree with the reference, i.e. Dev > tolerance.
- (atom name): Correct assignment, if within the same residue (no residue number given), or the neighboring residues.
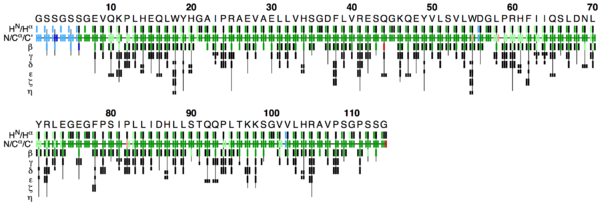
The flya.pdf file
This PDF file provides a graphical representation of the 'flya.tab' file. Each assignment for an atom is represented by a colored rectangle.
- Green: Assignment by FLYA agrees with the manually determined reference assignment (within tolerance)
- Red: Assignment by FLYA does not agree with the manually determined reference assignment
- Blue: Assigned by FLYA but no reference available
- Black: With reference assignment but not assigned by FLYA.
Respective light colors indicate assignments not classified as strong by the chemical shift consolidation. The row labeled HN/Hα shows for each residue HN on the left and Hα in the center. The N/Cα/C’ row shows for each residue the N, Cα, and C’ assignments from left to right. The rows β-η show the side-chain assignments for the heavy atoms in the center and hydrogen atoms to the left and right. In the case of branched side-chains, the corresponding row is split into an upper part for one branch and a lower part for the other branch.
FLYA applications
CYANA macros 'CALC*.cya' are provided for the following FLYA tasks:
CALC.cya: standard automated chemical shift assignment
- specify list of input peak lists in variable 'peaks' without intervening blanks
- specify tolerances for 1H, 13C, 15N with variables assigncs_assH, assigncs_assC assigncs_assN
- command 'select_atoms' excludes some nuclei that are difficult to detect
- optional parameter 'shiftreference=ref.prot' specifies reference chemical shift list, used only for comparison in flya.tab, flya.txt, flya.pdf
CALCbackbone.cya: standard backbone chemical shift assignment
- parameter 'structure=' to avoid generation of random structures, which are not needed if using only through-bond spectra
CALCexperiments.cya: using modified/new experiment definitions in library
- modified HCCHTOCSY only for aromatics (library HCCHTOCSY.lib, peak list HCCHTOCSYaro.peaks)
- new experiment N15NOESY2D (library peak list N15NOESY2D.lib, peak list N15NOESY2D.peaks)
CALCexpfromlist.cya: read expected peaks from a peak list
- command N15NOESY_expect, reading input peak list N15NOESY_in.peaks
CALCfixedpeaks.cya: keep input peak assignments in user peak assignments
- (partially) assigned input peak list N15HSQCassigned.peaks
- parameter 'keepassigned' for loadspectra.cya
CALCfixedshifts.cya: fix input chemical shift assignments
- input chemical shift list 'fix.prot'
- shift error in chemical shift list specifies range for assignment
CALClabeling.cya: use of experiment-specific isotope labeling
- command 'select_atoms' for general selection of assignable nuclei CcoNH + HSQCLEULYS
- command '<peak list name>_select' with atom selection for a specific peak list (e.g. C13HSQC_LK.peaks)
- command '<peak list name>_expect' for non-standard generation of expected peaks for a given peak list (e.g. CcoNH_LK.peaks with dimension-specific atom selection)
CALCnoesyonly.cya: chemical shift assignment using exclusively NOESY
- increased population size with 'shiftassign_population=200'
- see Schmidt et al. J. Biomol. NMR 57, 193-204 (2013)
CALCstatistics.cya: user-defined chemical shift statistics instead of standard BMRB statistics from library
- average value and stddev from input chemical shift list 'shiftx.prot'
- 'assigncs_sd:=bmrb' to use stddev from BMRB (cyana.lib) instead of input chemical shift list
- 'assigncs_sdfactor:=0.5' to scale BMRB stddev by given factor
CALCstructcalc.cya: follow automated shift assignment by automated NOESY assignment and structure calculation
- peak lists for distance restraint generation specified by parameter 'structurepeaks='
CALCstructure.cya: use input structure to generate expected peaks for through-space experiments
- specify with parameter 'structure' of command 'flya'
- if parameter 'structure' is absent, a set of random structures is generated automatically
- if set to blank ('structure='), no random structures are generated (if not needed because only through-bond spectra are used)
Results
You can download the results of all CYANA demo calculations (92 MB).