Basic structure calculation starting from given restraints: Difference between revisions
No edit summary |
No edit summary |
||
| Line 25: | Line 25: | ||
The first two commands read the input files with upper distance bounds and dihedral angle restraints, respectively. Then, 50 conformers are calculated using the standard [[simulated annealing]] schedule with 4000 [[torsion angle dynamics]] steps per conformer. Finally, the 10 conformers with the lowest final target function values are analyzed. An overview table of these 10 best conformers is saved in the file '''demo.ovw''', and their coordinates are written to the PDB file '''demo.pdb'''. | The first two commands read the input files with upper distance bounds and dihedral angle restraints, respectively. Then, 50 conformers are calculated using the standard [[simulated annealing]] schedule with 4000 [[torsion angle dynamics]] steps per conformer. Finally, the 10 conformers with the lowest final target function values are analyzed. An overview table of these 10 best conformers is saved in the file '''demo.ovw''', and their coordinates are written to the PDB file '''demo.pdb'''. | ||
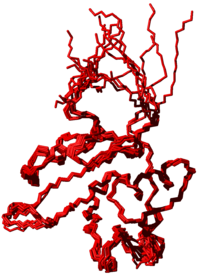
[[Image:StructuresDemoBasic.png|Structures obtained in a basic structure calculation starting from given restraints]] | [[Image:StructuresDemoBasic.png|thumb|200px|Structures obtained in a basic structure calculation starting from given restraints]] | ||
Revision as of 15:01, 2 June 2009
This calculation shows how to calculate a structure from previously assigned upper limits. The input data are in the subdirectory ‘basic’ of the demo data zip archive:
- demo.seq
- amino acid sequence
- demo.upl
- upper distance limit restraints
- demo.aco
- dihedral angle restraints
- init.cya
- initialization macro
- CALC.cya
- macro for the structure calculation
When CYANA is started, it automatically executes the commands in the initialization macro file init.cya, which usually holds basic setup for the calculations, e.g., reading the amino acid library and the amino acid sequence of the protein, and specifying general parameters such as the residue range for RMSD calculations. These commands could be put into the CALC.cya macro file (or even given on the command line), but as they are rarely changed for a given project, it is convenient have them in the init.cya file:
rmsdrange:=10-100 # define residue range for RMSD calculations cyanalib # read standard CYANA residue library read seq demo.seq # read amino acid sequence
The actual structure calculation is performed by the commands in the macro file CALC.cya:
read upl demo.upl # read upper distance bounds read aco demo.aco # read dihedral angle restraints calc_all 50 steps=4000 # calculate conformers overview demo.ovw structures=10 pdb # analyze and write best conformers
The first two commands read the input files with upper distance bounds and dihedral angle restraints, respectively. Then, 50 conformers are calculated using the standard simulated annealing schedule with 4000 torsion angle dynamics steps per conformer. Finally, the 10 conformers with the lowest final target function values are analyzed. An overview table of these 10 best conformers is saved in the file demo.ovw, and their coordinates are written to the PDB file demo.pdb.