Structure calculation with automated NOESY assignment: Difference between revisions
No edit summary |
No edit summary |
||
| (6 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
This calculation shows how to calculate a structure from previously unassigned NOESY peak lists. The input data are in the subdirectory | This calculation shows how to calculate a structure from previously unassigned NOESY peak lists. The input data are in the subdirectory ‘demo/auto’ of the CYANA software package: | ||
;demo.seq:amino acid sequence | ;demo.seq:amino acid sequence | ||
| Line 7: | Line 7: | ||
;demo.prot:<sup>1</sup>H, <sup>13</sup>C, and <sup>15</sup>N chemical shift list | ;demo.prot:<sup>1</sup>H, <sup>13</sup>C, and <sup>15</sup>N chemical shift list | ||
;demo.aco:dihedral angle restraints | ;demo.aco:dihedral angle restraints | ||
; | ;CALC.cya:macro for the structure calculation | ||
Combined automated NOESY cross peak assignment and structure calculation are performed with the macro file ''' | Combined automated NOESY cross peak assignment and structure calculation are performed with the macro file '''CALC.cya''': | ||
peaks := c13.peaks,n15.peaks,aro.peaks | peaks := c13.peaks, n15.peaks, aro.peaks # NOESY peak lists in XEASY format | ||
prot := demo.prot # names of chemical shift list(s) | |||
prot := demo.prot | restraints := demo.aco # additional (non-NOE) restraints | ||
restraints := demo.aco | tolerance := 0.04, 0.03, 0.45 # shift tolerances: H, H', C/N', C/N | ||
tolerance := 0. | #calibration_constant:=6.7E5,8.2E5,8.0E4 # calibration constants, automatic if empty | ||
#calibration_constant:=6.7E5,8.2E5,8.0E4 | structures := 100,20 # number of initial, final structures | ||
structures := 100,20 | steps := 10000 # number of torsion angle dynamics steps | ||
steps := 10000 | randomseed := 434726 # random number generator seed | ||
randomseed := 434726 | |||
noeassign peaks=$peaks prot=$prot autoaco | noeassign peaks="$peaks" prot="$prot" autoaco | ||
First several variables are set: The variable '''peaks''' gives the names of the input peak lists, separated by commas without intervening blanks. The variable '''prot''' gives the name(s) of the input chemical shift list(s). If a single name is given as in the example, it specifies that a single chemical shift list file with this name will be used for all peak lists. Alternatively, it is possible to specify a separate chemical shift list for each peak list as a comma-separated list of file names. The variable '''restraints''' specifies the names of input files with additional conformational restraints that will be used together with the upper distance bounds that will be derived from the NOESY peaks. If there are several file names, they must be separated by commas without intervening blanks. The variable [[tolerance]] specifies the tolerances for the matching of chemical shifts. It is used for a consistency check of the peaks that have assignments in the input peak lists, and for the automated NOESY cross peak assignment. The calibration constants for the peak lists can be given by the variable '''calibration''' as a comma-separated list of values in the order of the peak list names given by the variable '''peaks'''. If the variable '''calibration''' is not given, the calibration parameters are determined automatically such that the median of the upper distance limits for each peak list equals the value of the variable '''calibration_dref'''. The variable '''calibration_dref''' can have a single value that applies to all peak lists, or separate values for each peak list. This variable is not used when the calibration constants are given explicitly by the variable '''calibration_constant'''. | First several variables are set: The variable '''peaks''' gives the names of the input peak lists, separated by commas without intervening blanks. The variable '''prot''' gives the name(s) of the input chemical shift list(s). If a single name is given as in the example, it specifies that a single chemical shift list file with this name will be used for all peak lists. Alternatively, it is possible to specify a separate chemical shift list for each peak list as a comma-separated list of file names. The variable '''restraints''' specifies the names of input files with additional conformational restraints that will be used together with the upper distance bounds that will be derived from the NOESY peaks. If there are several file names, they must be separated by commas without intervening blanks. The variable [[tolerance]] specifies the tolerances for the matching of chemical shifts. It is used for a consistency check of the peaks that have assignments in the input peak lists, and for the automated NOESY cross peak assignment. The calibration constants for the peak lists can be given by the variable '''calibration''' as a comma-separated list of values in the order of the peak list names given by the variable '''peaks'''. If the variable '''calibration''' is not given, the calibration parameters are determined automatically such that the median of the upper distance limits for each peak list equals the value of the variable '''calibration_dref'''. The variable '''calibration_dref''' can have a single value that applies to all peak lists, or separate values for each peak list. This variable is not used when the calibration constants are given explicitly by the variable '''calibration_constant'''. | ||
[[Image:StructuresDemoManual.png|thumb|200px|'''demo.pdb''']] | [[Image:StructuresDemoManual.png|thumb|200px|'''demo.pdb''']] | ||
Seven cycles of combined automated NOESY assignment and structure calculation | Seven cycles of combined automated NOESY assignment and structure calculation, followed by a final structure calculation, are performed by the [[CYANA Macro: noeassign|noeassign]] macro, which uses the [[CYANA Command: assign|assign]] command to perform the NOESY assignment. In each cycle and in the final structure calculation 100 conformers are calculated using the standard [[simulated annealing]] schedule with 10000 [[torsion angle dynamics]] steps per conformer. The 20 conformers with the lowest final target function values are analyzed. An [[CYANA Macro: overview|overview]] table of these 20 best conformers is saved in the file '''final.ovw''', and their coordinates are written to the PDB file '''final.pdb'''. The corresponding files from the intermediate cycles 1-7 are called '''cycle1.*''', '''cycle2.*''', etc. | ||
An overview table of the complete calculation can be obtained with the command [[CYANA script: cyanatable|'''cyanatable''']] during or after the completion of the calculation. | An overview table of the complete calculation can be obtained with the command [[CYANA script: cyanatable|'''cyanatable''']] (at the Unix prompt) during or after the completion of the calculation. | ||
Latest revision as of 15:27, 28 July 2019
This calculation shows how to calculate a structure from previously unassigned NOESY peak lists. The input data are in the subdirectory ‘demo/auto’ of the CYANA software package:
- demo.seq
- amino acid sequence
- c13.peaks
- peak list from 3D 13C-resolved NOESY spectrum
- n15.peaks
- peak list from 3D 15N-resolved NOESY spectrum
- aro.peaks
- peak list from 3D aromatic 13C-resolved NOESY spectrum
- demo.prot
- 1H, 13C, and 15N chemical shift list
- demo.aco
- dihedral angle restraints
- CALC.cya
- macro for the structure calculation
Combined automated NOESY cross peak assignment and structure calculation are performed with the macro file CALC.cya:
peaks := c13.peaks, n15.peaks, aro.peaks # NOESY peak lists in XEASY format prot := demo.prot # names of chemical shift list(s) restraints := demo.aco # additional (non-NOE) restraints tolerance := 0.04, 0.03, 0.45 # shift tolerances: H, H', C/N', C/N #calibration_constant:=6.7E5,8.2E5,8.0E4 # calibration constants, automatic if empty structures := 100,20 # number of initial, final structures steps := 10000 # number of torsion angle dynamics steps randomseed := 434726 # random number generator seed noeassign peaks="$peaks" prot="$prot" autoaco
First several variables are set: The variable peaks gives the names of the input peak lists, separated by commas without intervening blanks. The variable prot gives the name(s) of the input chemical shift list(s). If a single name is given as in the example, it specifies that a single chemical shift list file with this name will be used for all peak lists. Alternatively, it is possible to specify a separate chemical shift list for each peak list as a comma-separated list of file names. The variable restraints specifies the names of input files with additional conformational restraints that will be used together with the upper distance bounds that will be derived from the NOESY peaks. If there are several file names, they must be separated by commas without intervening blanks. The variable tolerance specifies the tolerances for the matching of chemical shifts. It is used for a consistency check of the peaks that have assignments in the input peak lists, and for the automated NOESY cross peak assignment. The calibration constants for the peak lists can be given by the variable calibration as a comma-separated list of values in the order of the peak list names given by the variable peaks. If the variable calibration is not given, the calibration parameters are determined automatically such that the median of the upper distance limits for each peak list equals the value of the variable calibration_dref. The variable calibration_dref can have a single value that applies to all peak lists, or separate values for each peak list. This variable is not used when the calibration constants are given explicitly by the variable calibration_constant.
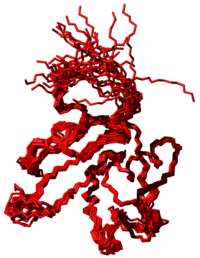
Seven cycles of combined automated NOESY assignment and structure calculation, followed by a final structure calculation, are performed by the noeassign macro, which uses the assign command to perform the NOESY assignment. In each cycle and in the final structure calculation 100 conformers are calculated using the standard simulated annealing schedule with 10000 torsion angle dynamics steps per conformer. The 20 conformers with the lowest final target function values are analyzed. An overview table of these 20 best conformers is saved in the file final.ovw, and their coordinates are written to the PDB file final.pdb. The corresponding files from the intermediate cycles 1-7 are called cycle1.*, cycle2.*, etc.
An overview table of the complete calculation can be obtained with the command cyanatable (at the Unix prompt) during or after the completion of the calculation.