Coordinate generation
Tree structure
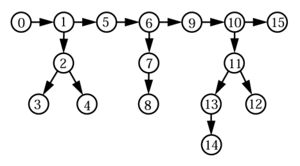
Structure calculations with the program CYANA work in torsion angle space. Since the degrees of freedom are torsion angles but distance restraints are defined by the Cartesian coordinates of the atoms, the program has to recompute the positions of the atoms after each change of the conformation, i.e. each change of the torsion angle values, in order to compute the target function. To this end, the molecule is considered as a tree, consisting of rigid units that are connected by rotatable bonds (Abe et al., 1984; Güntert, 1993; Güntert et al., 1991). Rigid units do not contain rotatable bonds and are made up by one or several atoms whose relative positions are invariable.
The torsion angles and the rigid units are numbered with monotonously increasing numbers, if the tree is traversed in the direction of the arrows, which point in the direction of that part of the tree that is moved by a rotation about a rotatable bond. A rigid unit and the (always unique) rotatable bond that leads to it have the same number a and uniquely defined predecessors p(a) < a. A rigid unit a comprises all atoms, whose position depends only on the torsion angle a, if all preceding torsion angles a’ < a are held fixed. For instance, in Fig. 1 the rigid units 1, 6 and 9 each comprise the atoms Hα, C’ and Cβ, and the rigid units 5 and 9 both comprise the atoms O, N, H and Cα (the peptide bond is not rotatable).
Torsion angles
A torsion angle a is defined by four atoms with position vectors , , and . The rotatable bond connects the second with the third atom of the torsion angle definition. The second atom of the torsion angle definition is the start point of the rotatable bond. The unit vector along the axis of the rotatable bond is given by . The value ϕa ∈ [-π,π] of the torsion angle a is related to the position vectors , , and by
- ,
where and , and ϕa receives the same sign as ea. The symbols “⋅” and “Λ” denote scalar and the vector product, respectively.
Coordinate generation
Upon reading the residue library, each individual residue is rotated (as a whole) into a standard orientation:
The position vector of the atom α = 1,2,… in the residue library is denoted by rα. The corresponding position vector in the standard orientation of the residue is . R(u,v)is the rotation matrix whose rows are given by the three orthonormal vectors
- , and .
To calculate the coordinates of the atoms in a conformation that is given by the torsion angle values (ϕ1,…,ϕn) residues can be attached to a partial polypeptide chain one by one. Afterwards – before appending the next residue – the required torsion angle values must be set by rotating the appropriate atoms of the new residue. The “overlap atoms” in the residue library are used to dock a residue to the preceding one. The first three atoms of each residue entry in the residue library (the atoms C, O and N for amino acid residues in the standard residue library) are superimposed onto the final three atoms with corresponding names of the preceding residue. The coordinates of the atoms of the docked residue are obtained by the transformation
- ,
where p1, p2 and p3 are the positions of the overlap atoms of the preceding residue, and are the coordinates of the new residue in standard orientation defined above. The transformation
- (α ∈ Ma).
sets the desired torsion angle values. The transformation has to be applied for all rotatable bonds m of the newly docked residue in descending order, m,m - 1,…,1. In this equation ra denotes the position vector of the start point of the rotatable bond a, and ea is a unit vector along the rotation axis of the rotatable bond a. The desired torsion angle value is given by ϕa, and is the value of the torsion angle a in the conformation whose coordinates are stored in the residue library. Ma denotes the set of atoms that are moved when the torsion angle a is rotated. R(e,ϕ) is the matrix for a rotation with axis e and rotation angle ϕ, given by
for an arbitrary vector x.
The generation of the atom coordinates using the above equations is inefficient because an atom is in general subjected to several coordinate transformations. The program CYANA therefore uses these equations only once after reading the residue sequence to generate the atom coordinates of the conformation with torsion angle values ϕ1 = … = ϕn = 0. This conformation is the start point for the fast algorithm that is used to generate the atom coordinates during torsion angle dynamics and minimization by building the “scaffold” of the rotatable bonds followed by the positioning of the atoms by a single coordinate transformation. Starting from the conformation ϕ1 = … = ϕn = 0 with atom coordinates {r&alpha}, the rigid units are first brought into a standard orientation:
- .
denotes the position vector of the ith atom in the definition of the torsion angle a. The transformation is applied to all atoms α in the rigid unit a. In addition, for each rigid unit a a rotation matrix Ra and a translation vector ta are calculated – the two quantities that form the scaffold of the tree structure.
- ,
- .
The quantities , Ra and ta have to be computed only once after reading the amino acid sequence. For each new conformation (ϕ1,…,ϕn)
- (ez = (0,0,1))
Builds recursively the scaffold of rotatable bonds, and
generates the coordinates with a single transformation for each atom.
References
ABE, H., BRAUN, W., NOGUTI, T. & GO, N. (1984). Rapid calculation of 1st and 2nd derivatives of conformational energy with respect to dihedral angles for proteins - General recurrent equations. Computers & Chemistry 8, 239–247.
GÜNTERT, P. (1993). Neue Rechenverfahren für die Proteinstrukturbestimmung mit Hilfe der magnetischen Kernspinresonanz. Ph.D. thesis, ETH.
GÜNTERT, P., BRAUN, W. & WÜTHRICH, K. (1991). Efficient computation of three-dimensional protein structures in solution from nuclear magnetic resonance data using the program DIANA and the supporting programs CALIBA, HABAS and GLOMSA. Journal of Molecular Biology 217, 517–530.